Gradient Boosting
Table of Contents
Gradient boosting
Overview
Gradient boosting is a technique for regression and classification problems, which produces a prediction model in the form of an ensemble of weak prediction models, typically prediction trees. It builds the model in a stage-wise fashion like other boosting methods do, and generalizes them by allowing optimization of an arbitrary differentiable loss function.
Motivation
Consider a least-squares regression setting, where the goal is to "teach" a model  to predict values in the form
to predict values in the form  by minimizing the mean squared error
by minimizing the mean squared error  , averaged over some training set of actualy values of the output variable
, averaged over some training set of actualy values of the output variable 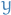 .
.
At each stage  , of gradient boosting, it may be assumed that there is a some imperfect model
, of gradient boosting, it may be assumed that there is a some imperfect model  . The gradient boosing algorithm improves on
. The gradient boosing algorithm improves on  by constructing a new model that adds an estimator
by constructing a new model that adds an estimator  to provide a better model:
to provide a better model:  .
.
To find  , the gradient boosting solution starts with the observation that a perfect
, the gradient boosting solution starts with the observation that a perfect  would imply
would imply

i.e. that adding the extra predictor  will make up for all the mistakes our previous model
will make up for all the mistakes our previous model  did, or equivalently
did, or equivalently

Therefore, gradient boosting will fit  to the residual
to the residual  . Like in other boosting variants, each
. Like in other boosting variants, each  learns to correct its predecessor
learns to correct its predecessor  .
.
The generalization to other loss-functions than the least-squares, comes from the observation that  for a given model are the negative gradients (wrt.
for a given model are the negative gradients (wrt.  ) of the squared error loss function, hence for any loss function
) of the squared error loss function, hence for any loss function  we'll simply fit to the negative gradient
we'll simply fit to the negative gradient  .
.
Algorithm
Input: training set  , a differentiable loss function
, a differentiable loss function  , number of iterations
, number of iterations 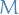 .
.
Initialize model with a constant value:
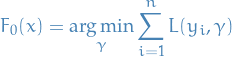
For
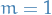 to
to 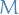 :
:
- Compute so-called psuedo-residuals :
![\begin{equation*}
r_{im} = - \Bigg[ \frac{\partial L(y_i, F(x_i))}{\partial F(x_i)} \Bigg]_{F(x) = F_{m-1}(x)} \quad \text{for } i = 1, \dots, n
\end{equation*}](../../assets/latex/boosting_bf5ab350fd8f28957deb4ae6d333d2aee3bd9983.png)
- Fit a base learner (e.g. tree)
 to psuedo-residuals, i.e. train it using the training set
to psuedo-residuals, i.e. train it using the training set 
- Compute multiplier
 sby solving the following one-dimension optimization problem
sby solving the following one-dimension optimization problem

- Update the model:

- Output

Gradient Tree Boosting
- Uses decision trees of fixed size as base learners
Modified gradient boosting method ("TreeBoost")
- Generic gradient boosting at the m-th step would fit a decision tree
 to psuedo-residuals
to psuedo-residuals - Let
 be the number of its leaves
be the number of its leaves Tree partitions input space into
 disjoint regions
disjoint regions  and predicts a constant value in each region, i.e.
and predicts a constant value in each region, i.e.
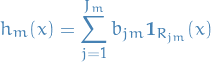
where
 is the value predicted in the region
is the value predicted in the region  .
.
In the case of usual CART trees, where the trees are fitted using least-squares loss, and so the coefficient  for the region
for the region  is equal to just the value of the output variable, averaged over all training instances in
is equal to just the value of the output variable, averaged over all training instances in  .
.
Continuing with generic gradient boosting, the model is:
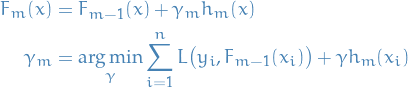
The modified gradient tree boosting ("TreeBoost") algorithm is then to instead choose a separate optimal value  for each of the tree's regions, instead of a single
for each of the tree's regions, instead of a single  for whole tree:
for whole tree:
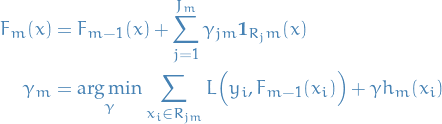
where we've simply absorbed  into
into  .
.
Terminology
- Out-of-bag error / estimate is a method for measuring prediction error of methods utilizing bootstrap aggregating (bagging) to sub-sample data used for training.
OOB is the mean prediction error on each training sample
 , using only the trees that did not have
, using only the trees that did not have  in their bootstrap sample
in their bootstrap sample
Regularization
- Number of trees: Large number of trees can overfit
Shrinkage: use a modified update rule:

where
 is the learnin rate.
is the learnin rate.
- Subsampling is when at each iteration of the algorithm, a base learner should be fit on a subsample of the training set drawn at random without replacement. Sumsample size
 is the fraction of training set to use; if
is the fraction of training set to use; if  then algorithm is deterministic and identitical to TreeBoost
then algorithm is deterministic and identitical to TreeBoost - Limit number of observations in trees' terminal nodes: in the tree building process by ignoring any split that lead to nodes containing fewer than this number of training set instances. Helps reduce variance in predictions at leaves.
- Penalize complexity of tree: model complexity can be defined as proportional number of leaves in the learned trees. Joint optimization of loss and model complexity corresponds to a post-pruning algorithm.
- Regularization (e.g.
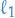 ,
, 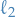 ) on leaf nodes
) on leaf nodes